MANTA basic
MANTA (Microbiota And pheNotype correlaTion Analysis platform) is a software program, and MANTA can create an integrative database and analysis platform that relates microbiome and phenotypic data. It is a web application which works on most of the modern web browsers.
MANTA basic is developed for personal usage. Instead of processing the data and then import them into the database directly, we implement a data manage function, that the data could be uploaded via the user interface in the web application. On the other hand, some tables were deprecated in order to simplify the upload process. The following sections describe how this data management function works.
Get the application
MANTA basic (desktop version) for Windows and MacOS is ready-to-use.
- For Windows user: After download the file, please unzip it and execute LaunchManta.exe.
- For Mac user: After download the file, please run the installer.
Data format
There are 2 type of files; 'microbiota' and 'phenotype parameters'.
Microbiota
The currently accepted microbiota file format is a tab-delimited table format (see the sample data as an example), which can be easily converted from the BIOM (The Biological Observation Matrix) format file (https://biom-format.org/). The BIOM format is adopted by several popular projects including QIIME and MG-RAST. Kraken/Bracken produces results in a different format and we are working on enabling the system to import this type of data.
Phenotype parameters
The accepted format is a tab delimited table format that contains different parameters in each column and different samples in each row.
The sample data set can be download form here. (same as the 20 samples in the MDD)
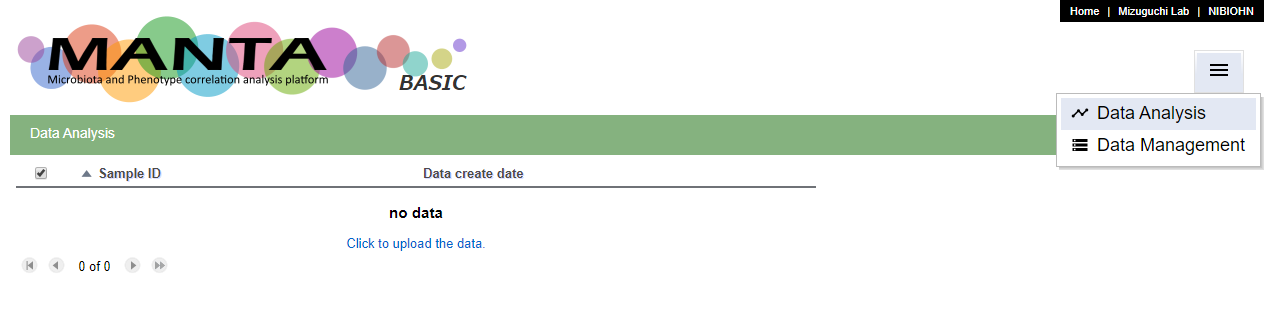
Data management
You can upload your data from the 'Data management' function. You can access it from the menu at upper right or if there is no data in the database, there is a link 'Click to upload the data' in the empty table.
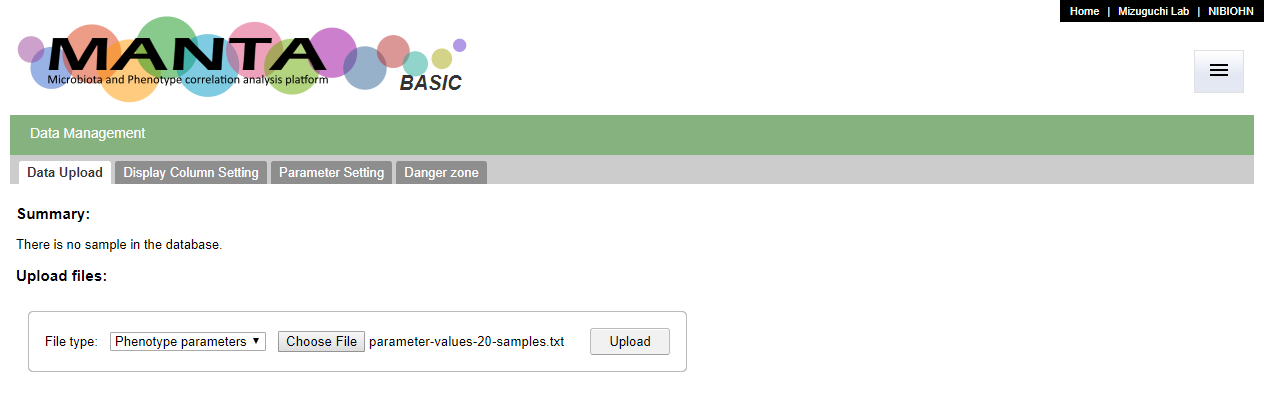
Upload parameters
To upload the (phenotypic) parameter data, select 'Phenotype parameters' as the 'File type', and choose the data file. The file should be a tab delimited file. The first column should be the sample identifiers (ids) and the first row should be the header of different parameters.
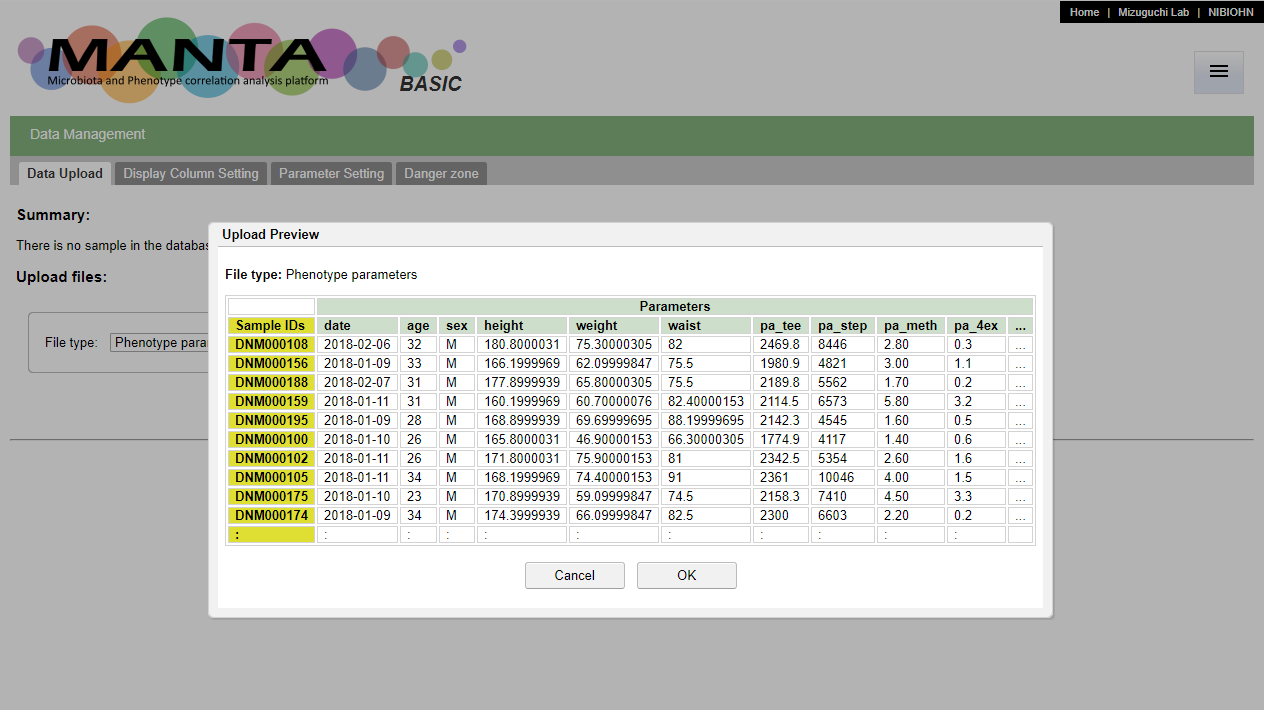
After click on the 'Upload' button, there is a dialog window showing the preview of the file. If the sample ids and the parameter header are allocated correctly, press 'OK' to finish data upload.
Upload microbiota
To upload the microbiome data, select 'Microbiota' as the 'File type', and choose the data file. The file should be a tab delimited file. The first column should be the the hierachy of taxonomy from kindom (e.g. Bacteria), separated by the semicolumn (;). The first row should be the header of sample identifiers (ids).
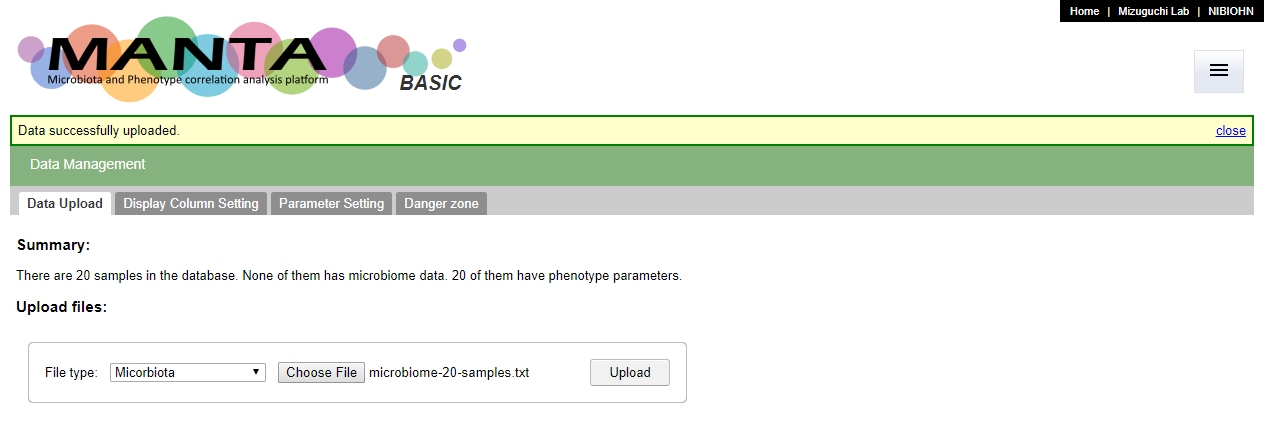
After click on the 'Upload' button, there is a dialog window showing the preview of the file. If everything is fine, press 'OK' to finish data upload.
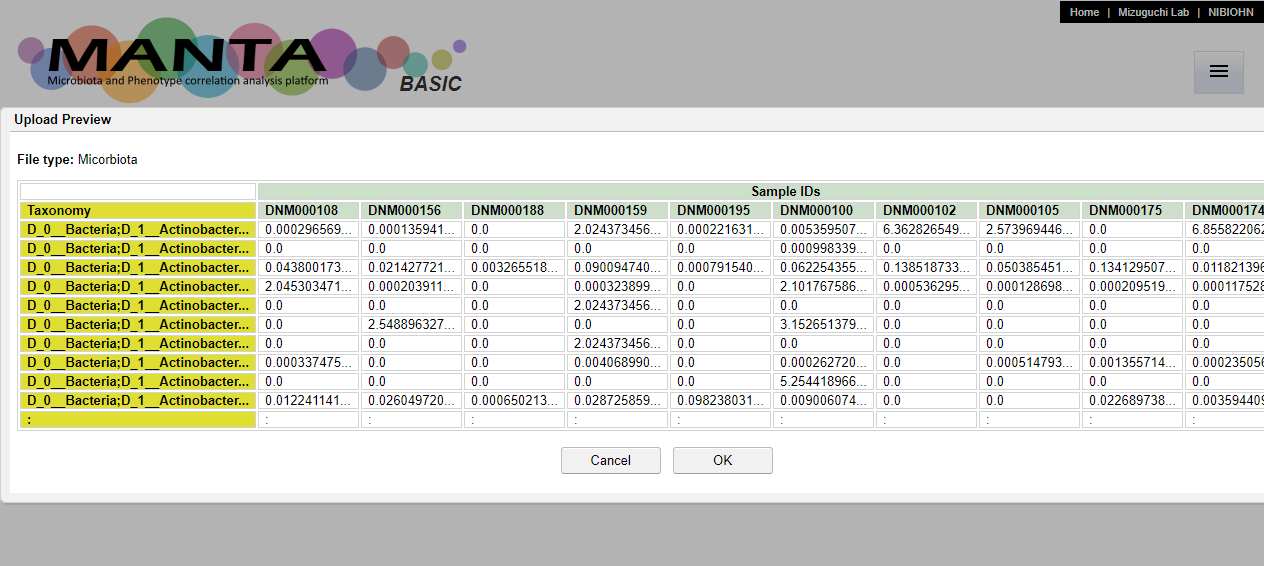
Set display columns
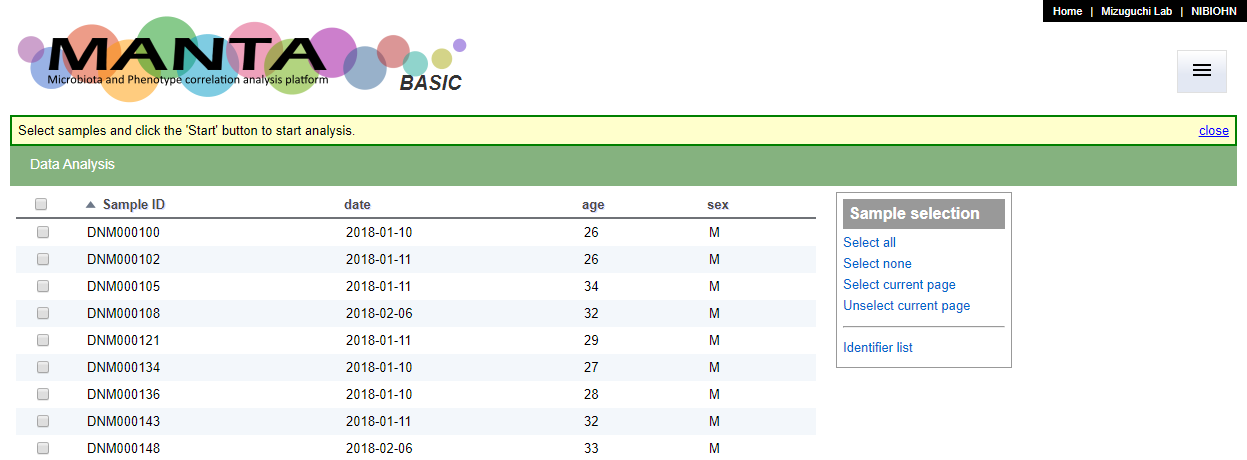
By default, the main page (Data Analysis page), will only show a list of sample identifiers (see the screen shot below).
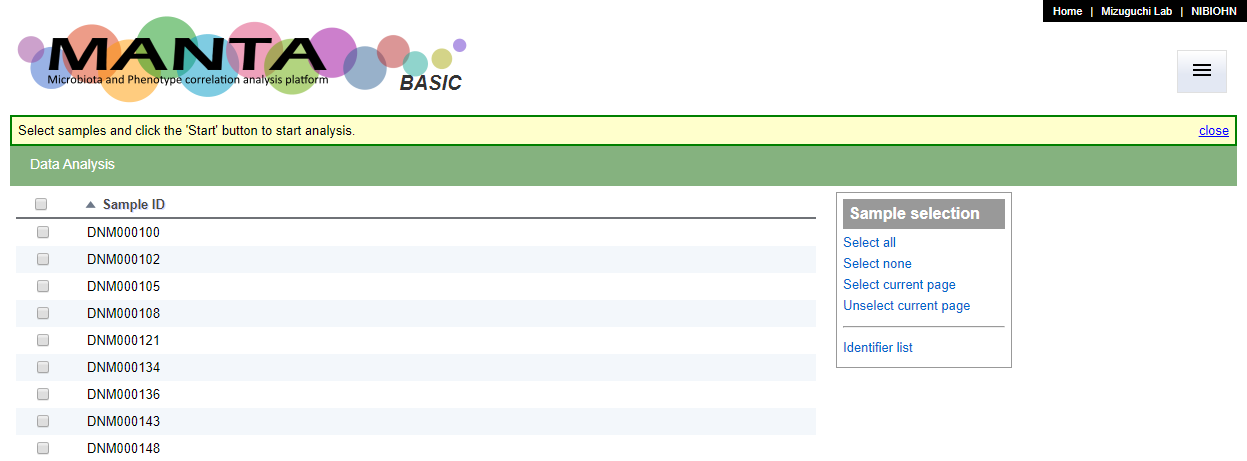
The display columns could be configurate in the 'Display Column Setting' tab in the 'Data Management' function. You can choose upto 3 uploaded parameters to display.
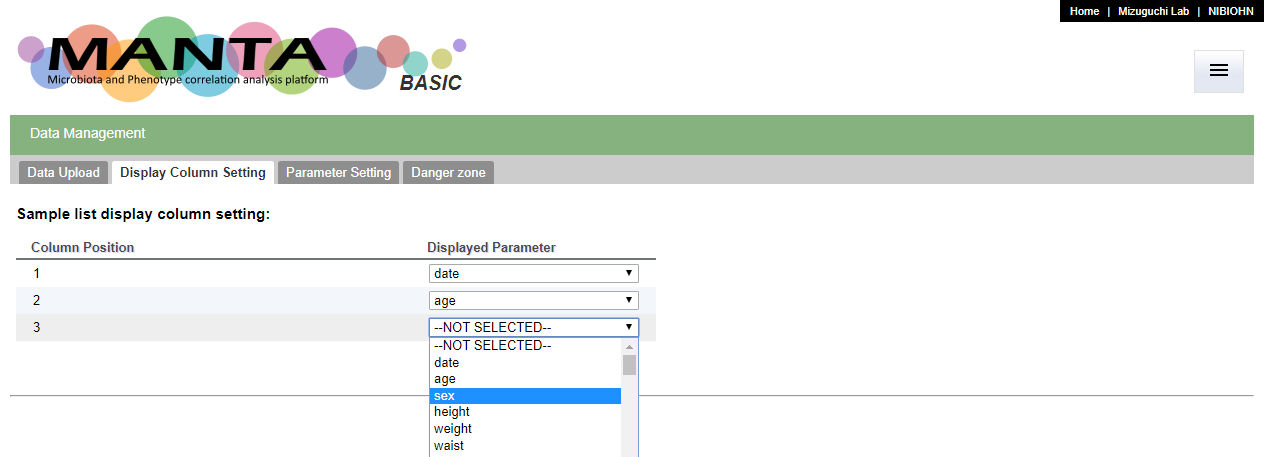
The selected parameters will be displayed in the sample list table (as below).
Set parameter type
By default, all the uploaded parameters are set to the 'free text' type. You can set the parameter types in the 'Parameter Setting' tab in the 'Data Management' function.
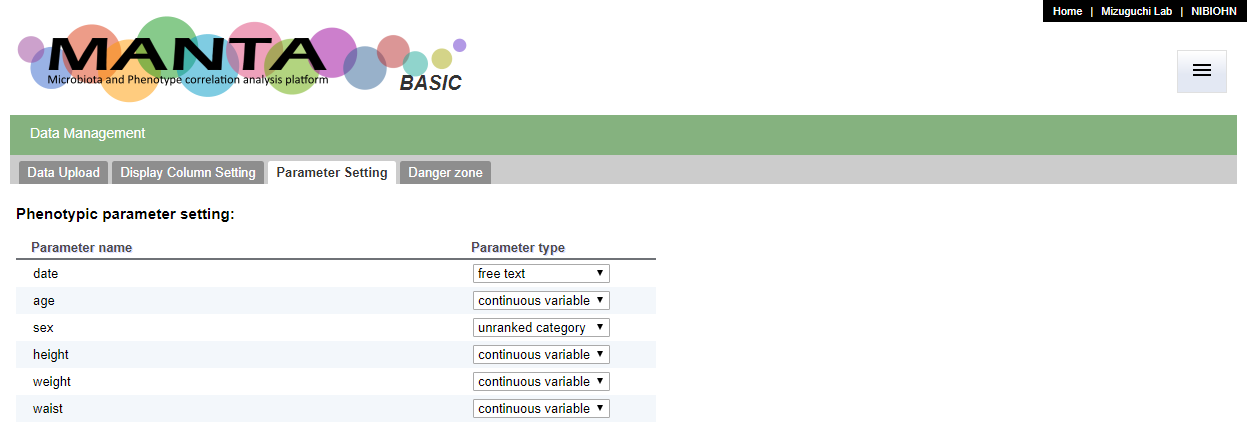
Only the 'continuous variable' type could be used for calculating the correlation coefficient. Only the 'continuous variable', 'unranked category' and 'ranked category' could be used to color the data points in the PCoA plot. The 'others' type was designed for the data types that do not belong to any of the rest. At the moment, this type of data is treated in the same way as the type 'free text', and not used for any correlation calculation.
We are working on the auto assignment of the data types. This feature will be added in a future release.
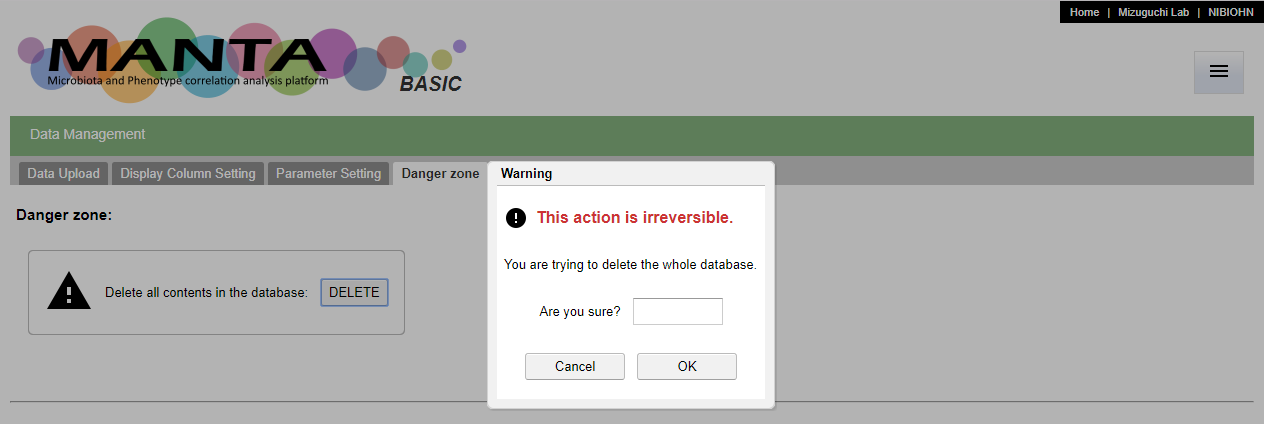
Delete database
It is possible to clear the whole database via the user interface. You can find the 'DELETE' button in the 'Danger zone' tab in the 'Data Management' function. This action is irreverible. Please be careful to use this function.
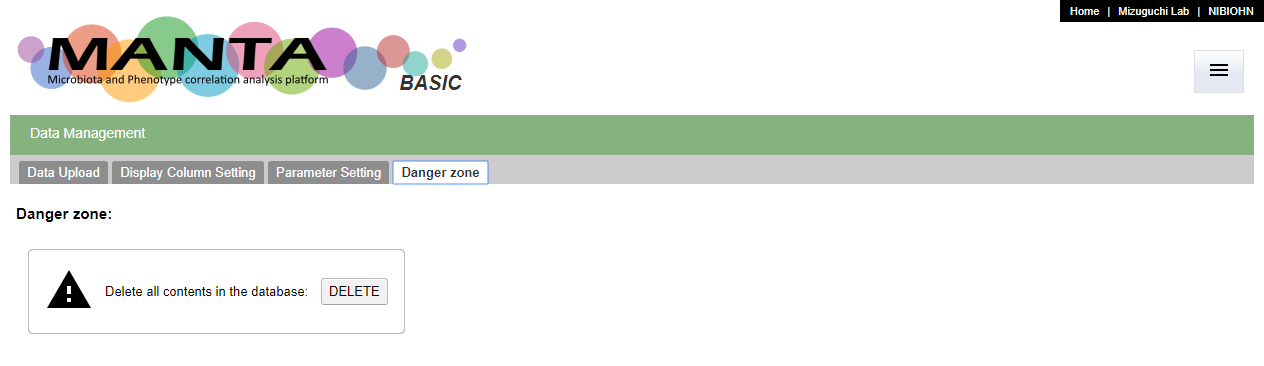
This action will remove all data in the database, including all the settings for the parameters. Again, this action is irreverible. You will need to re-import the data and do all settings again if you would like to redo the analysis. Type 'yes' or 'y' to confirm the operation.